Table 1. Primer selection table for specific 16S rRNA gene region to be amplified.
| Region Covered | Primer Selection | Primer Sequence (5′-3′) | Amplicon Size (bp) | Reference |
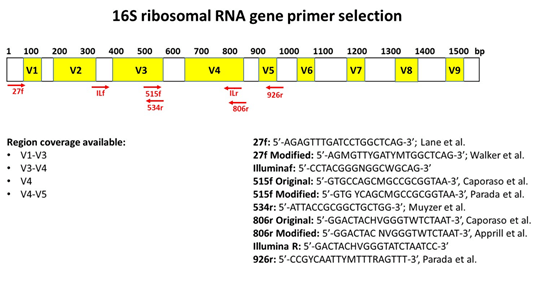
| V1 – V3 | 27F | AGAGTTTGATCCTGGCTCAG | 507 | 5 |
| 27F (modified) | AGMGTTYGATYMYGGCTCAG | 6 | ||
| 534R | ATTACCGCGGCTGCTGG | 7 | ||
| V3 – V4 | IlluminaF | CCTACGGGGNGGCWGCAG | 465 | 1 |
| IlluminaR | GACTACHVGGGTATCTAATCC | 1 | ||
| V4 | 515F (modified) | GTGCCAGCMGCCGCGGTAA | 291 | 2 |
| 515F (modified) | GTGYCAGCMGCCGCGGTAA | 3 | ||
| 806R | GGACTACHVGGGTWTCTAAT | 2 | ||
| 806R (modified) | GGACTACNVGGGTWTCTAA | 4 | ||
| V4 -V5 | 515F (modified) | GTGCCAGCMGCCGCGGTAA | 411 | 2 |
| 515F (modified) | GTGYCAGCMGCCGCGGTAA | 3 | ||
| 926R | GGACTACHVGGGTWTCTAAT | 3 |
Note: Illumina adapter sequences that are added to primer (not included in amplicon size in Table 1):
Forward adapter: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGâ€[locus specific sequence]
Reverse adapter: 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGâ€[locus specific sequence]
Figure 1. Gene diagram showing relative position of primers in relation to variable regions.
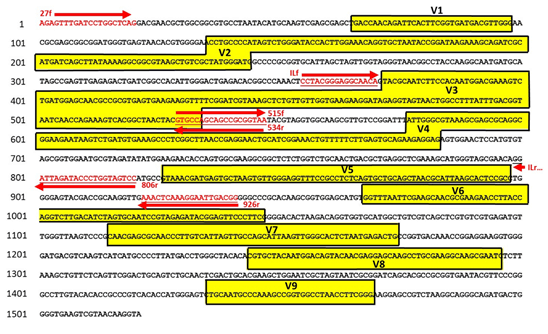
Figure 2. Ribosomal RNA gene sequence of Lactobacillus acidophilus (EF533992.1) with putative primer binding sites and variable regions shown.
References:
- http://support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html
- Caporaso JG,et al. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 108 (Suppl):4516–4522. doi:10.1073/pnas.1000080107.
- Parada AE, et al. 2015. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. doi:10.1111/1462-2920.13023.
- Apprill A, et al. 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 75:129–137. doi:10.3354/ame01753.
- Jiang, H., et al. 2006. Microbial Diversity in Water and Sediment of Lake Chaka, an Athalassohaline Lake in Northwestern China. Applied and Environmental Microbiology. 72 (6): 3832-3845. doi:10.1128/AEM.02869-05.
- Walker, A.W., et al. 2015. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3:26. doi: 10.1186/s40168-015-0087-4.
- Muyzer, G. et al. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:3 695-700.